Abstract
Background: Pediatric immune thrombocytopenia purpura (ITP) is the most common cause of autoimmune cytopenias in children and results in autoimmune destruction of platelets, leading to significantly decreased circulating platelets, increased bleeding risk and fatigue. The underlying mechanism of action is thought to be auto-antibody driven, though auto-reactive T cells have also been implicated in some cases. Given the proximity of many cases of ITP to either recent viral infection or vaccine administration, a leading hypothesis is that the environmental exposure mimics a platelet produced epitope causing platelet clearance. Despite the correlation of viral infection and/or inflammatory cascades in the propagation of ITP, the gut microbiome in newly diagnosed pediatric patients has not previously been interrogated. The gut microbiome has previously been shown to modulate the host inflammatory milieu and dysbiosis has been associated with other auto-immune disorders. Thus, we assessed the gut microbiome in newly diagnosed pediatric ITP patients to determine if a similar dysbiosis is present.
Methods: Stool samples were collected from 32 pediatric patients (0-18 years) at the Children's Hospital of Philadelphia. 17 patients were newly diagnosed with ITP, 7 patients had ITP for greater than 3 months at time of stool collection and 8 patients had newly diagnosed severe acquired aplastic anemia. Stool samples were kept on ice until processing at our PENN/CHOP Microbiome Core (not longer than 24 hours/sample). Age matched healthy control samples were provided by the Microbiome Core. Shotgun libraries were generated from 0.5 ng DNA using the Nextera XT Library Prep kit and libraries were sequenced on an Illumina HiSeq 2500 in High Output mode to produce paired-end 125 bp sequence reads. Shotgun metagenomic data were analyzed using Sunbeam, a user-extendable bioinformatics pipeline that we developed for this purpose.Diversity within samples were assessed by the number of OTUs at a rarefaction level of 1,000 sequences and the Shannon index. Sample similarity were assessed by Bray-Curtis and Jaccard distances, which were then visualized using principle coordinates analysis.
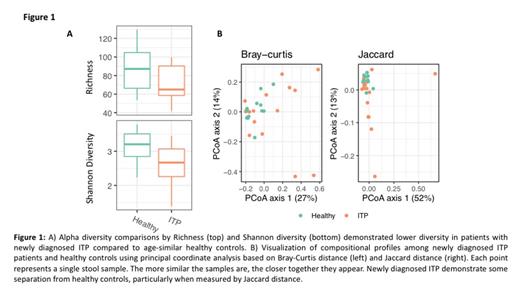
Results: Subjects included 17 patients with newly diagnosed ITP (i.e. stool sample collected < 3 months from diagnosis), 7 patients with ITP > 3 months and 8 patients with newly diagnosed severe acquired aplastic anemia. Patient samples were assessed for gut microbial diversity and richness. As seen in figure 1a, gut microbial diversity in newly diagnosed pediatric patients with ITP was significantly decreased (p = 0.024) while gut microbial richness trended to a concomitant decrease as well (p = 0.093). Interestingly, healthy controls had similar gut microbiomes to one another than newly diagnosed ITP patients had to one another (Fig 1b). These alterations in the gut microbiome were not seen in pediatric patients with newly diagnosed aplastic anemia or with ITP diagnosed > 3 months prior to collection (data not shown). These findings indicate that while there is a dysbiotic gut microbiome in patients with newly diagnosed ITP, there is no dysbiosis present in other pediatric autoimmune cytopenias. Furthermore, the dysbiosis corrects by 3 months post-ITP diagnosis. When we assess specific bacterial families and subgroups we do not find a predominant species that is increased or decreased but rather there are global changes amongst many bacterial species. We did find a trend of lower Alistipes species and increased Eschrechia coli in newly diagnosed ITP patients which is similar to other autoimmune disorders with shifted gut microbiota. Functional pathways analyses showed similar global alterations in vital pathways such as decreased aminoacyl tRNA biosynthesis and homologous recombination.
Conclusion: Pediatric patients with newly diagnosed ITP but not other autoimmune cytopenias have gut microbial dysbiosis with perturbations in many bacterial species that may be a cause of, or a result of the underlying mechanism of pathogenesis. Further studies will help determine the role of dysbiosis in the pathobiology of ITP and determine whether intervention alters the duration of disease.
Lambert: Bayer: Consultancy; Astra Zeneca: Research Funding; Shionogi: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; ClinGen, ISTH, ASH, GW University: Honoraria; PDSA: Research Funding; Novartis: Consultancy, Honoraria, Research Funding; Rigel: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Sysmex: Research Funding; Principia: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Argenx: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Octapharma: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Dova: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal